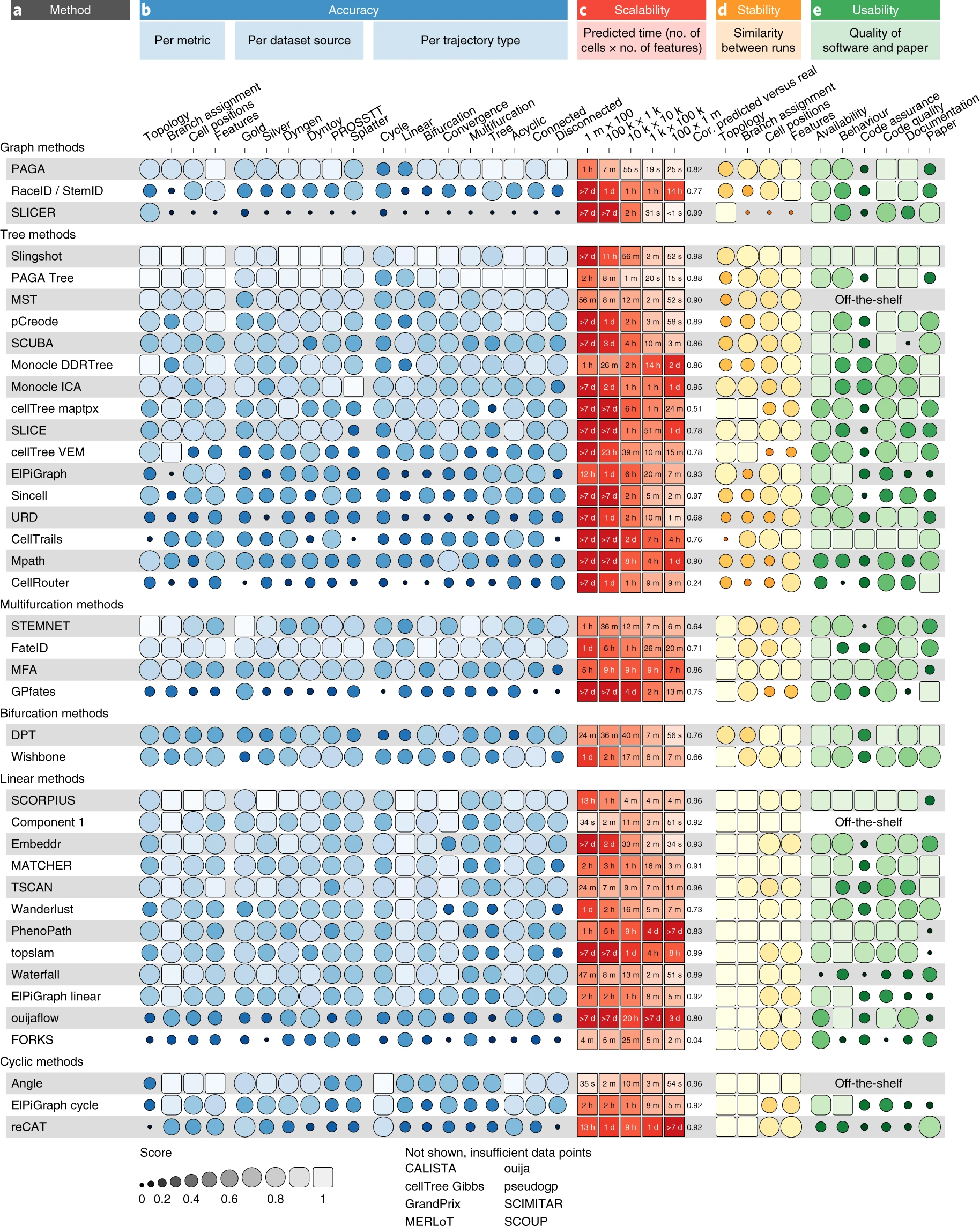
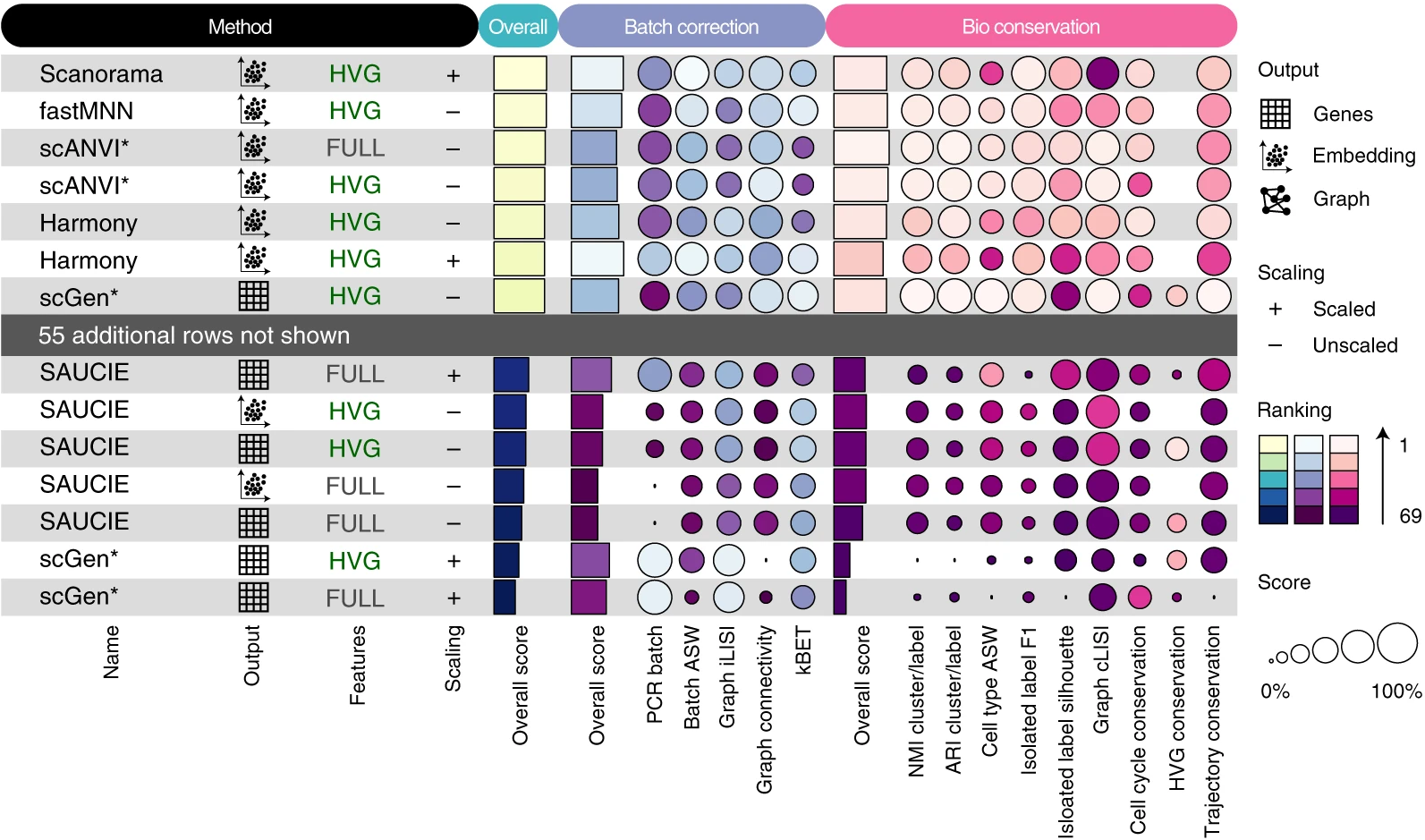
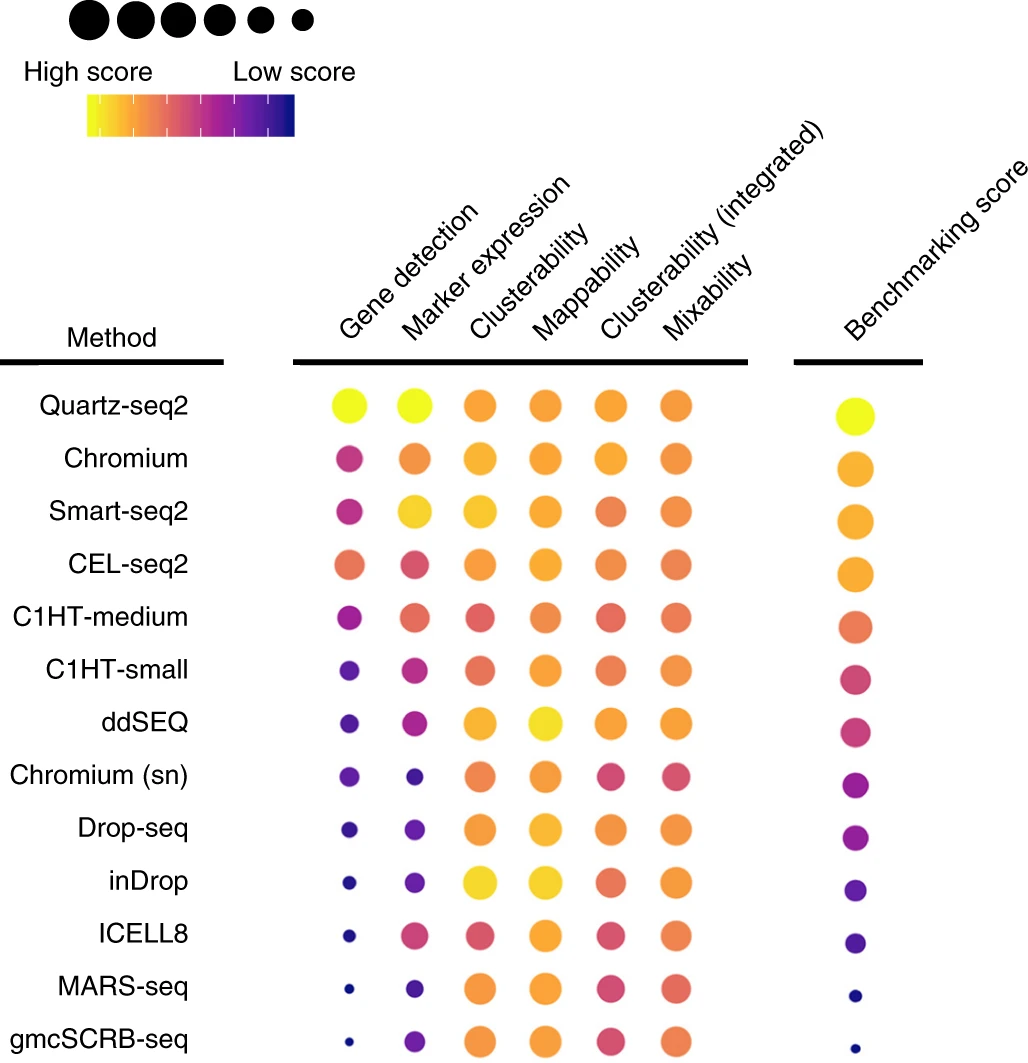
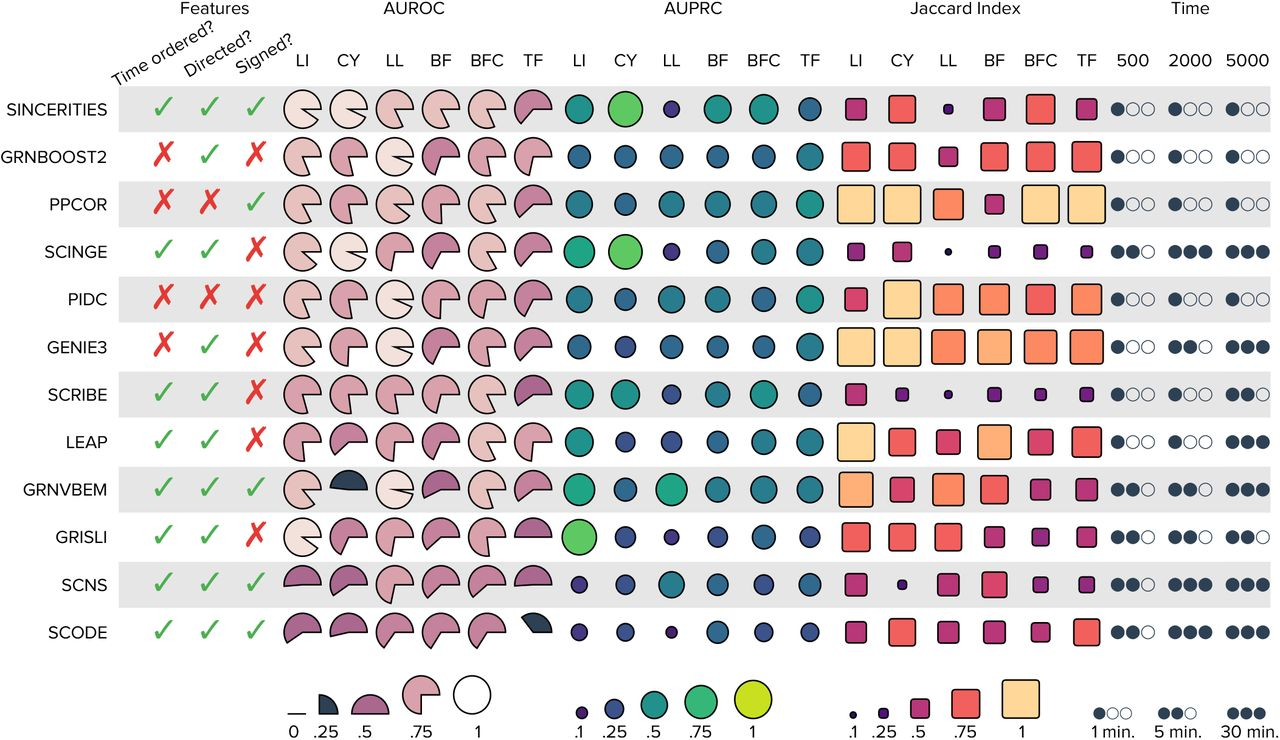
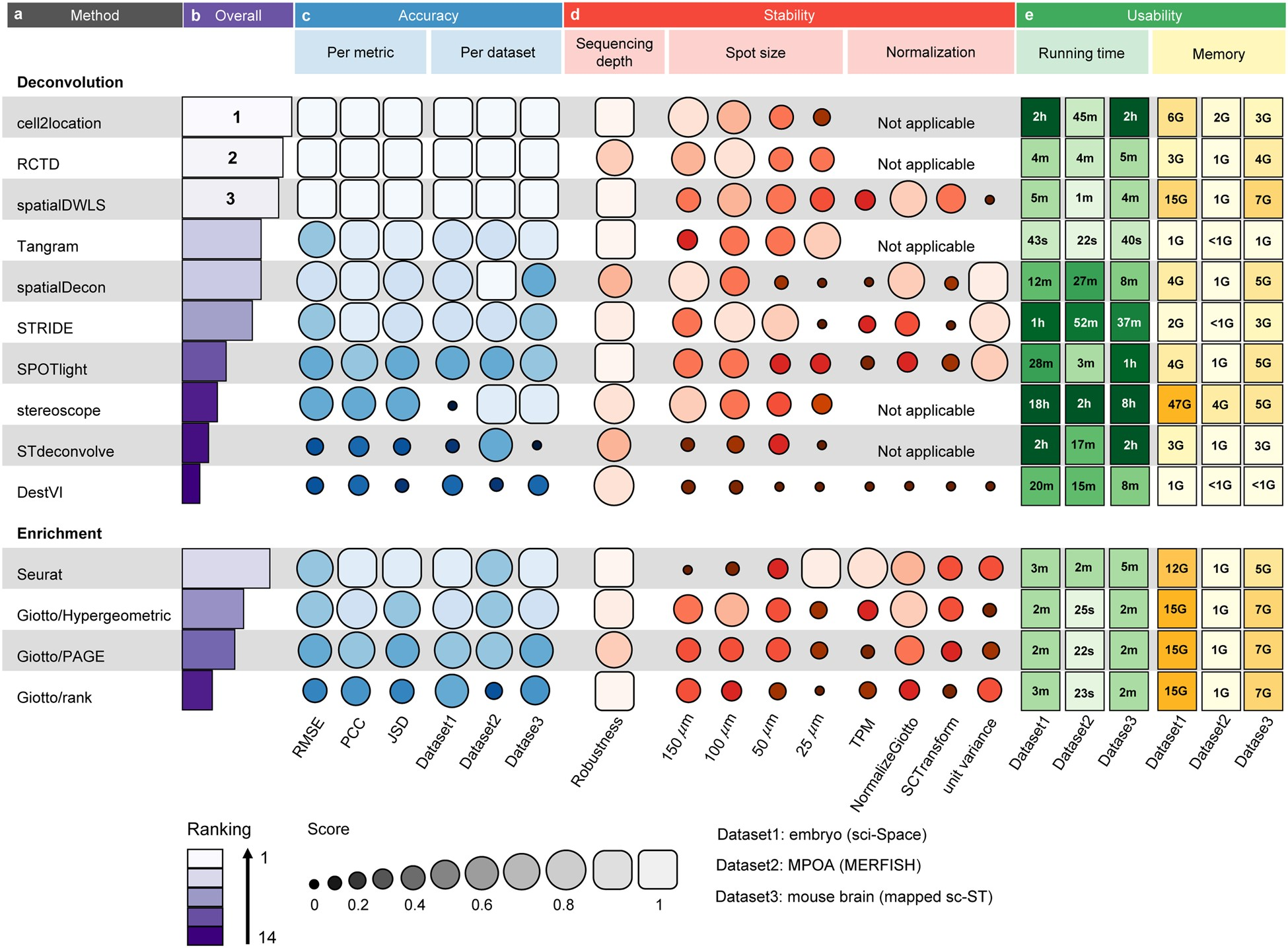
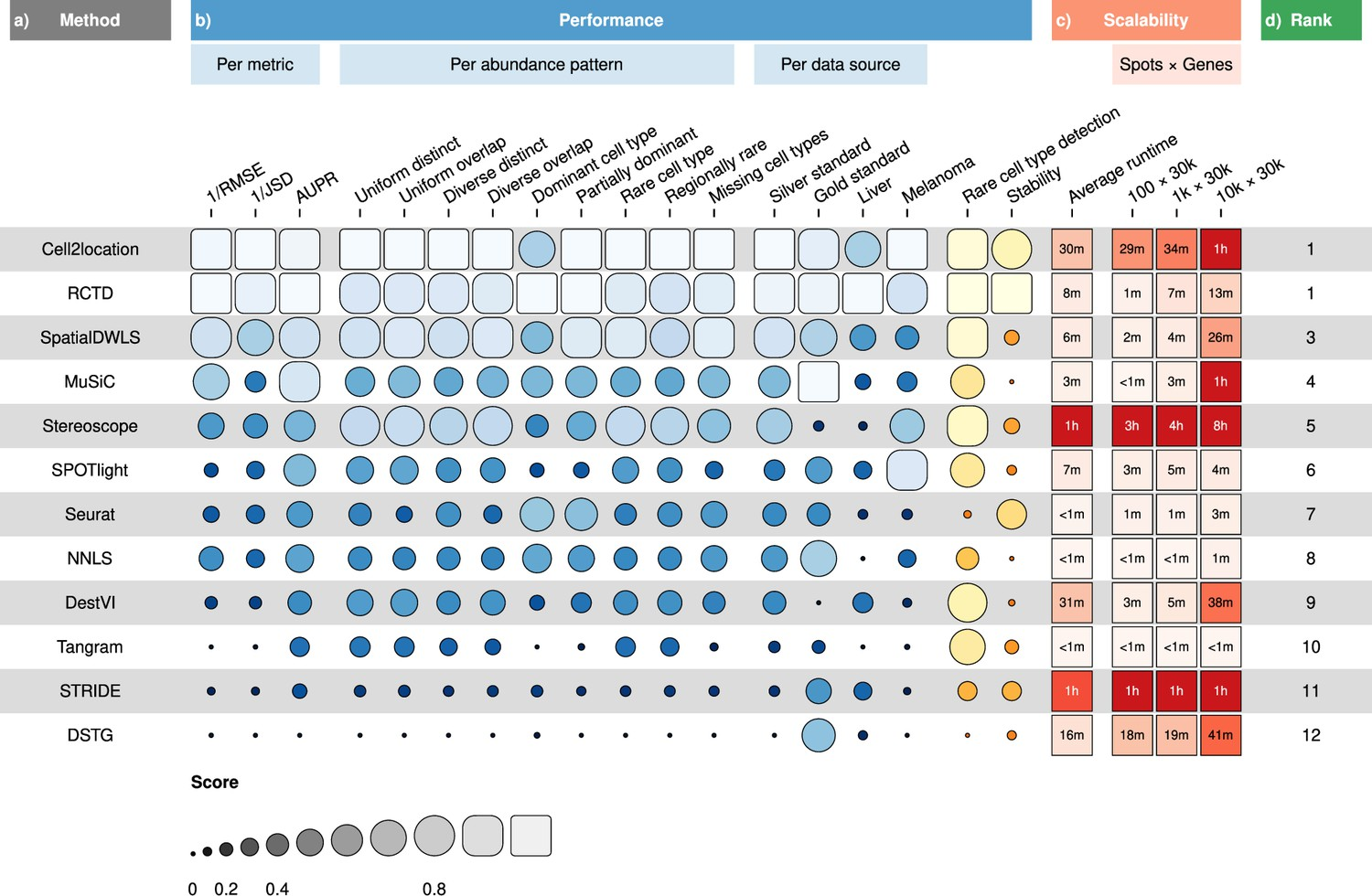
{funkyheatmap} allows generating heatmap-like visualisations for data frames. Funky heatmaps can be fine-tuned by providing annotations of the columns and rows, which allows assigning multiple palettes or geometries or grouping rows and columns together in categories.
Implementations
Gallery
These are works with funkyheatmap (-inspired) visualisations. Add your work to the list by creating a PR!